This class is a general MMSD Model. More...
#include <MMSD_Model.h>
Public Member Functions | |
| void | setIsEmptyClusterDeleted (const tBoolean &v) |
| if true the empty clusters are deleted during esperaceEvaluation() defalut value: false. see MMSD_Model:::esperanceEvaluation() More... | |
| void | setDataSet (SP::MMSD_DataSet data) |
| set the data set More... | |
| const MMSD_DataSet * | getDataSet () const |
| get the data set More... | |
| MMSD_DataSet * | getDataSet () |
| get the data set More... | |
| void | setEMMaximumIterationsNumber (const int &n) |
| set the maximum iterations number for EM algorithm More... | |
| int | getEMMaximumIterationsNumber () const |
| get the maximum iterations number for EM algorithm More... | |
| int | getEMIterationsNumber () const |
| get the number of iterations done More... | |
| tVectorIndex | getClustersNumber () const |
| get clusters number More... | |
| const MMSD_Cluster * | getCluster (const tVectorIndex &index) const |
| get the cluster at index More... | |
| const MMSD_DoubleFullMatrix & | getSampleClusterProbabilities () const |
| get the probability of samples to be in cluster matrix of size NxK More... | |
| const MMSD_IntegerVector & | getSampleClusterIndices () const |
| get the cluster of samples vector of size N More... | |
| void | setMinEigenValue (const double &v) |
| set min eigen value for eigen value decomposition of property covariance matrix law More... | |
| double | getMinEigenValue () const |
| get min eigen value for eigen value decomposition of property covariance matrix law More... | |
| virtual SP::MMSD_Cluster | NewClusterInstance () const =0 |
| create a cluster More... | |
| virtual SP::MMSD_Law | NewLawInstance () const =0 |
| create a cluster More... | |
| void | setWeightInitializationType (const tString &type, const double &scale, const double &rate) |
| set the weight initialization type & parameters More... | |
| tBoolean | parametersOptimizationByEMMethod (const int &nClusters, const MMSD_IntegerVector &sampleClusters, const tString &backupPath, const tString &backupPrefix, const int &nDigits, const int &backupSteps) |
| optimise the parameters of each laws by EM algorithm More... | |
| tBoolean | parametersOptimizationByEMMethod (const int &nClusters, const MMSD_IntegerVector &sampleClusters) |
| optimise the parameters of each laws by EM algorithm More... | |
| tBoolean | parametersOptimizationByEMMethod () |
| optimise the parameters of each laws by EM algorithm More... | |
| tBoolean | parametersOptimizationByEMMethod (const tString &backupPath, const tString &backupPrefix, const int &nDigits, const int &backupSteps) |
| optimise the parameters of each laws by EM algorithm More... | |
| tBoolean | restore (SP::CORE_ClassFactory factory, const tString &backupPath, const tString &backupPrefix, const int &nDigits, const int &step) |
| restore the model from a file More... | |
| tBoolean | restore (SP::CORE_ClassFactory factory, const tString &file) |
| restore the model from a backup file More... | |
| void | computeSampleClusterProbabilities (MMSD_DoubleFullMatrix &P, MMSD_DoubleVector &work) const |
| compute clusters probability for each sample More... | |
| void | computeSampleClusterProbabilities () |
| compute clusters probability for each sample More... | |
| void | computeSampleClusterIndices (MMSD_DoubleVector &work, MMSD_IntegerVector &indices) const |
| compute the index of cluster for each sample More... | |
| tBoolean | esperanceEvaluation () |
| compute Esperance Evaluation step More... | |
| tBoolean | esperanceMaximization () |
| compute Esperance Maximization step More... | |
| tBoolean | hasConverged () const |
| test of convergence More... | |
| tBoolean | computeLogLikelihood (double &logL) |
| compute logLikelihood More... | |
| const MMSD_DoubleVector & | getLogL () const |
| get logL for each iteration of the optimization process More... | |
| virtual void | saveToUIClass (UI_Class &mclass) const |
| how to save the object from a mate model class More... | |
| virtual void | loadFromUIClass (const UI_Class &mclass) |
| how to load the object from a Meta Model class More... | |
| virtual tString | toString () const |
| turn the class into string More... | |
| void | setHasBeenLoaded (const tBoolean &v) |
| set the if the object has completely been loaded More... | |
| tBoolean | hasBeenLoaded () const |
| return true if the object has completely been loaded More... | |
| void | getSharedPointer (SP::CORE_Object &p) |
| get the shared pointer of this class into p More... | |
| void | getSharedPointer (SPC::CORE_Object &p) const |
| get the shared pointer of this class into p More... | |
| tString | getClassName () const |
| return the class name of the object More... | |
| tString | getIdentityString () const |
| return the identity string of the object of the form className_at_address More... | |
| tString | getPointerAddress () const |
| return the identity string of the object More... | |
| template<class T > | |
| tBoolean | isInstanceOf () const |
| return true if the object is an instance of T More... | |
| virtual void | print () |
| print the class More... | |
| virtual ostream & | print (ostream &out) const |
| print the class More... | |
| virtual void | print (const tString &message) |
| print the class More... | |
| virtual void | print (const tInteger &str) |
| print More... | |
| virtual void | print (const tRelativeInteger &str) |
| print More... | |
| virtual void | print (const tReal &str) |
| print More... | |
| virtual void | print (const int &str) |
| print More... | |
Static Public Member Functions | |
| static void | initSeed (const long int &v) |
| init the seed with v value More... | |
| static void | initSeed () |
| init the seed More... | |
| static tString | getClassName (const tString &identityString) |
| return the class name of the object using only the identity string More... | |
| template<class T > | |
| static tString | getTypeName () |
| get type name More... | |
| static tBoolean | is64Architecture () |
| return true if the machine is a 64 bits machine More... | |
| static tBoolean | is32Architecture () |
| return true if the machine is a 32 bits machine More... | |
| static tString | pointer2String (const void *obj) |
| return the string represantation of a pointer More... | |
| static void | setOutput (ostream &out) |
| set output More... | |
| static ostream & | getOutput () |
| get output More... | |
| static void | printObjectsInMemory () |
| print object in memory More... | |
| static ostream & | print (ostream &out, const tString &message) |
| print the class More... | |
| static void | outputPrint (const tString &message) |
Static Public Attributes | |
| static tBoolean | mIsMemoryTesting =false |
| indicator to store all classes created and deleted only for debuging version More... | |
Protected Member Functions | |
| MMSD_Model (void) | |
| create an object More... | |
| virtual | ~MMSD_Model (void) |
| destroy an object. More... | |
| virtual void | restore (const MMSD_DoubleFullMatrix &properties) |
| restore the unstored values after a backup More... | |
| virtual void | initialize (const MMSD_IntegerVector &clusters, const MMSD_DoubleFullMatrix &properties, const int &nClusters) |
| initialization INPUT parameters: More... | |
| void | setThis (SP::CORE_Object p) |
| set this weak shared pointer called toDoAfterThis setting method More... | |
| virtual void | setType (tString type) |
| set the type of the object More... | |
| virtual void | toDoAfterThisSetting () |
| method called after setThis() method this method can oly be called once. More... | |
Detailed Description
This class is a general MMSD Model.
A model is a class which makes clustering of samples.
It needs:
- to define the model cluster MMSD_Model::NewClusterInstance() method
- to define the model law MMSD_Model::NewLawInstance() method
a MMSD_Model class has:
- a list of clusters MMSD_Cluster of size K
- a sample probability to be in cluster k in [0,K[ (matrix of size NxK) see (MMSD_Model::getSampleClusterProbabilities())
The main method of the class is MMSD_Model::parametersOptimizationByEMMethod().
The number of iterations of this EM Method is set by the method MMSD_Model::setEMMaximumIterationsNumber(). This method calls successivly MMSD_Model::esperanceEvaluation() && MMSD_Model::esperanceMaximization(). The value of the optimized function is computed in MMSD_Model::computeLogLikelihood() method.
The computing of sample probabilities to be in cluster is done by MMSD_Model::computeSampleClusterProbabilities() method which use the method MMSD_Cluster::computeMultivariateDensity()
The sample properties described by MMSD_DataSet are set by MMSD_Model::setDataSet()
- Version
- 1.0
Constructor & Destructor Documentation
|
protected |
|
protectedvirtual |
destroy an object.
Member Function Documentation
|
inline |
compute logLikelihood
- Parameters
-
logL OUTPUT value of log-likelihood function
- Returns
- true if the computing succeeds
Referenced by parametersOptimizationByEMMethod().

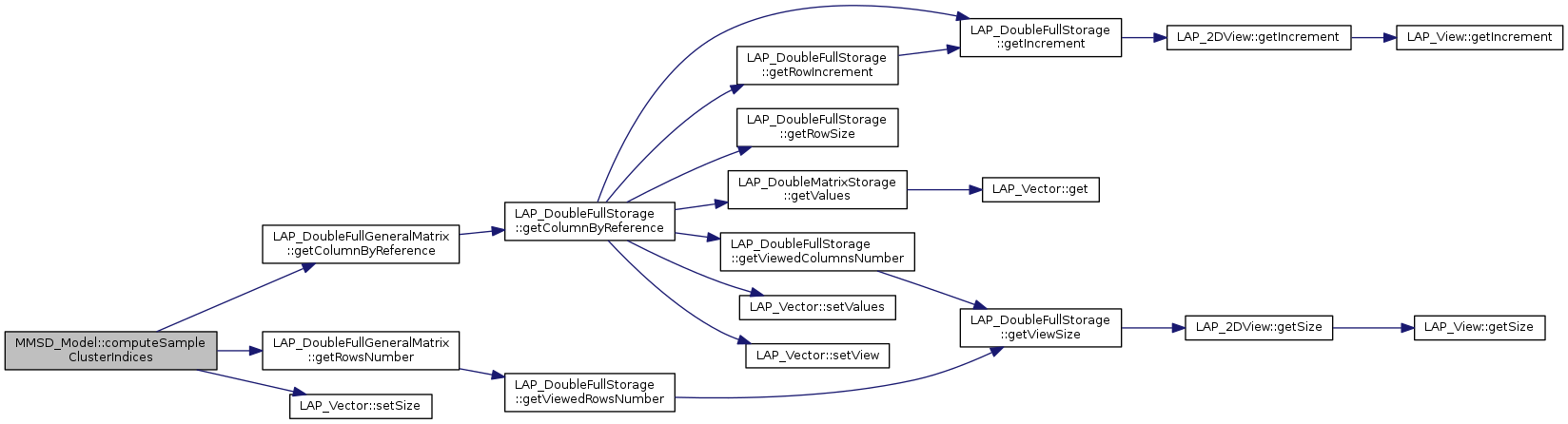
| void MMSD_Model::computeSampleClusterIndices | ( | MMSD_DoubleVector & | work, |
| MMSD_IntegerVector & | indices | ||
| ) | const |
compute the index of cluster for each sample
- Parameters
-
work is an array of working, used for memory optimization indices : output integer vector of storing the index of cluster for each sample.
indice[i]=index of cluster for with the probability Pk[i] is max
References LAP_DoubleFullGeneralMatrix::getColumnByReference(), LAP_DoubleFullGeneralMatrix::getRowsNumber(), and LAP_Vector< T >::setSize().
Referenced by parametersOptimizationByEMMethod().

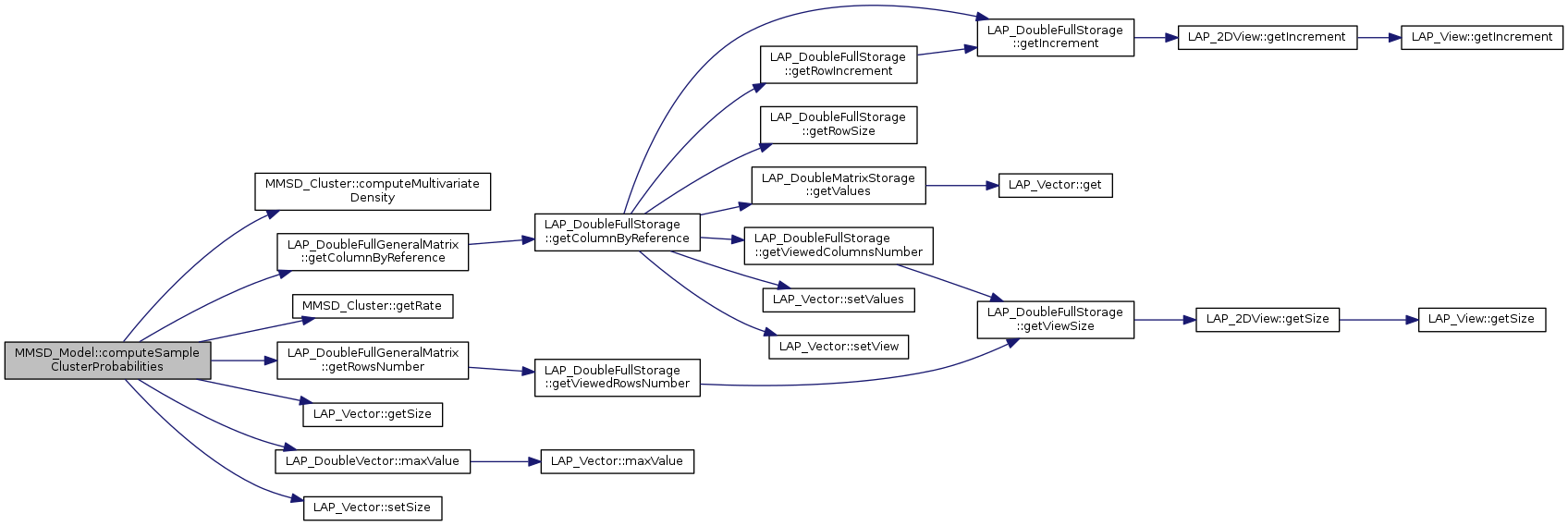
| void MMSD_Model::computeSampleClusterProbabilities | ( | MMSD_DoubleFullMatrix & | P, |
| MMSD_DoubleVector & | work | ||
| ) | const |
compute clusters probability for each sample
- Parameters
-
P OUTPUT parameter the cluster probability of each sample work array of size nThreads*nSamples
work array is used for temporary computing.
N is the number of samples, K is the number of clusters For each cluster k in [0,K[
- compute the multivariateDensity of the cluster Pk of size N (see MMSD_Cluster::computeMultivariateDensity())
- to each value of Pk , add log of cluster rate (see MMSD_Cluster::getRate())
- compute the max value of all Pk : Pmax
For each cluster k in [0,K[
- Pk:=exp(Pk-Pmax)
- compute the sum of the Pk: S[i]+=Pk[i] i in [0,N[;
For each cluster k in [0,K[
- Pk[i]/=S[i] i in [0,N[;
References MMSD_Cluster::computeMultivariateDensity(), LAP_DoubleFullGeneralMatrix::getColumnByReference(), MMSD_Cluster::getRate(), LAP_DoubleFullGeneralMatrix::getRowsNumber(), LAP_Vector< T >::getSize(), LAP_DoubleVector::maxValue(), LAP_Vector< T >::setSize(), and tLVectorIndex.
|
inline |
compute clusters probability for each sample
Referenced by esperanceMaximization(), initialize(), and restore().
| tBoolean MMSD_Model::esperanceEvaluation | ( | ) |
compute Esperance Evaluation step
for each cluster k in [0,K[
- get the probability of all samples to be in cluster k called Pk
- update the rate of the cluster k (see MMSD_Cluster::updateRate() )
- if the cluster is empy :
- if empty clusters are not deleted, raises an error (see MMSD_Model::setIsEmptyClusterDeleted() ) mIsEmp
- if not, erase it.
update the weights of cluster k (see MMSD_Cluster::updateWeights())
References LAP_DoubleFullGeneralMatrix::getColumnByReference(), LAP_Vector< T >::getValues(), MMSD_Cluster::isEmpty(), LAP_DoubleFullGeneralMatrix::removeColumn(), LAP_Vector< T >::setSize(), CORE_Integer::toString(), MMSD_Cluster::updateRate(), and MMSD_Cluster::updateWeights().
Referenced by parametersOptimizationByEMMethod().

| tBoolean MMSD_Model::esperanceMaximization | ( | ) |
compute Esperance Maximization step
- for each cluster in [0,K[
- get the probability of all samples to be in cluster k called Pk
- update the law of the cluster (see MMSD_Cluster::updateLaw()) with properties data set & Pk
- update the freedom degrees of the cluster (see MMSD_Cluster::updateFreedomDegrees()) with Pk
- sort the cluster parameters see MMSD_Cluster::sort()
- recompute the probability of all samples to be in cluster k See MMSD_Model::computeSampleClusterProbabilities()
References computeSampleClusterProbabilities(), LAP_DoubleFullGeneralMatrix::getColumnByReference(), MMSD_Cluster::sort(), MMSD_Cluster::updateFreedomDegrees(), and MMSD_Cluster::updateLaw().
Referenced by parametersOptimizationByEMMethod().

return the class name of the object using only the identity string
Referenced by UI_Class::createVector2D(), LAP_DoublePackedUpperMatrix::matrixProduct(), LAP_DoubleBandedUpperMatrix::matrixProduct(), LAP_DoubleFullUpperMatrix::matrixProduct(), and CORE_Object::printObjectsInMemory().
|
inherited |
return the class name of the object
- Returns
- the class name of the object
References tString.
Referenced by CORE_Object::getIdentityString().

|
inline |
get the cluster at index
References null.
Referenced by parametersOptimizationByEMMethod().
|
inline |
get clusters number
|
inline |
get the data set
Referenced by parametersOptimizationByEMMethod().

|
inline |
get the data set
|
inline |
get the number of iterations done
|
inline |
get the maximum iterations number for EM algorithm
|
inlineinherited |
return the identity string of the object of the form className_at_address
- Returns
- the identity string of the object
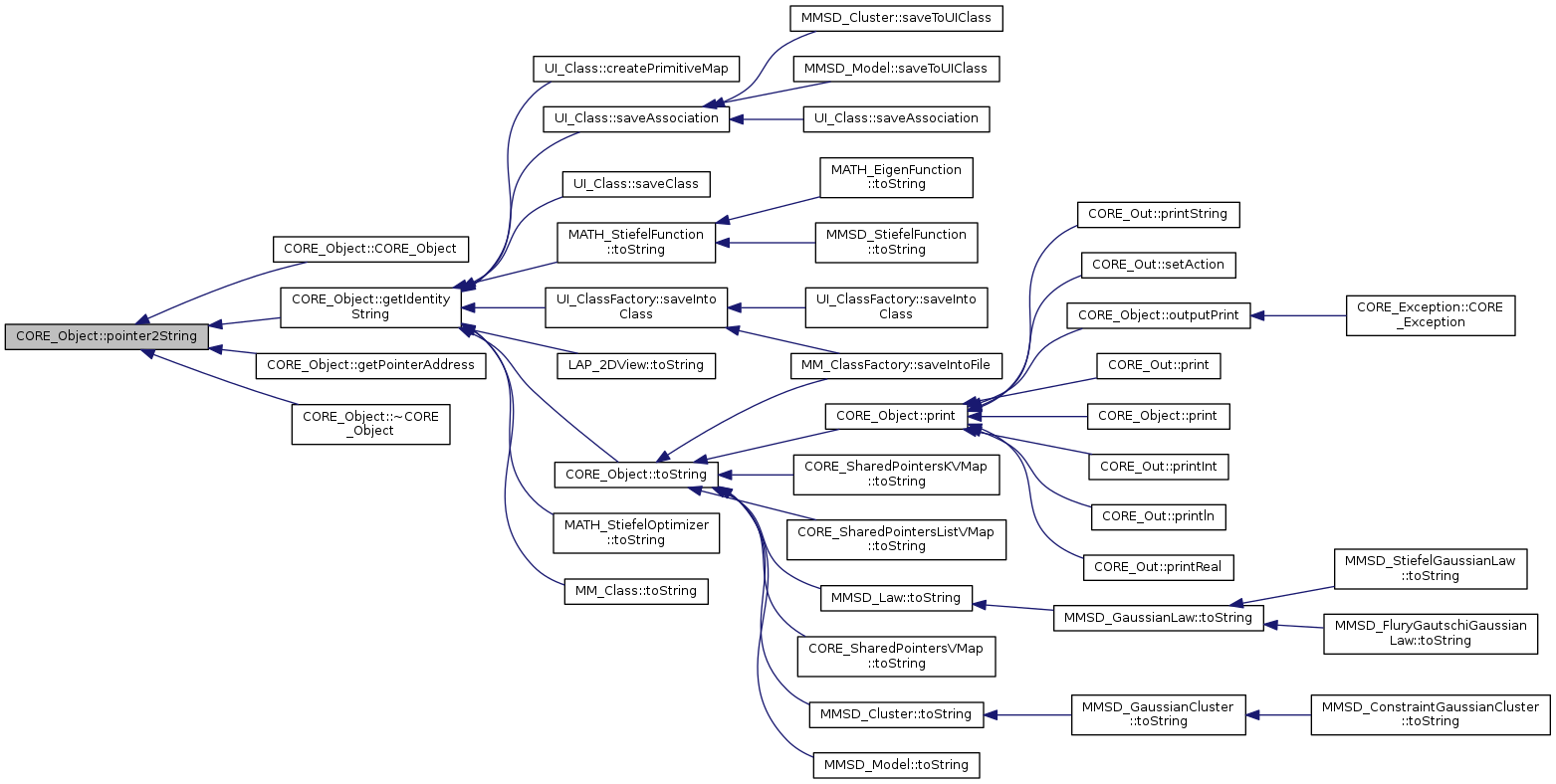
References CORE_Object::getClassName(), CORE_Object::pointer2String(), and tString.
Referenced by UI_Class::createPrimitiveMap(), UI_Class::saveAssociation(), UI_Class::saveClass(), UI_ClassFactory::saveIntoClass(), MATH_StiefelFunction::toString(), LAP_2DView::toString(), CORE_Object::toString(), MATH_StiefelOptimizer::toString(), and MM_Class::toString().

|
inline |
get logL for each iteration of the optimization process
|
inline |
get min eigen value for eigen value decomposition of property covariance matrix law
Referenced by MMSD_GaussianModel::NewLawInstance().
|
inlinestaticinherited |
get output
|
inlineinherited |
return the identity string of the object
- Returns
- the identity string of the object
References CORE_Object::pointer2String().

|
inline |
get the cluster of samples vector of size N
|
inline |
get the probability of samples to be in cluster matrix of size NxK
|
inlineinherited |
get the shared pointer of this class into p
Referenced by CORE_Map< Key, Value >::getSharedPointer(), CORE_Array< tFlag >::getSharedPointer(), LAP_ConstVector< lapack_real >::getSharedPointer(), STAT_Combinatorial< T >::getSharedPointer(), LAP_Vector< lapack_int >::getSharedPointer(), CORE_Vector< T >::getSharedPointer(), CORE_Matrix< T >::getSharedPointer(), and CORE_Object::printObjectsInMemory().

|
inlineinherited |
get the shared pointer of this class into p
|
inlinestaticinherited |
get type name
References tString.
|
inlineinherited |
return true if the object has completely been loaded
Referenced by UI_Class::loadAssociation().
| tBoolean MMSD_Model::hasConverged | ( | ) | const |
test of convergence
Referenced by parametersOptimizationByEMMethod().

|
protectedvirtual |
initialization INPUT parameters:
- Parameters
-
clusters : index of cluster for each sample properties properties matrix nClusters number of clusters
- Build the clusters list:
- creates a cluster MMSD_Model::NewClusterInstance()
- creates a law of the cluster MMSD_Model::NewClusterLaw()
- attachs the law to the cluster MMSD_Cluster::setLaw()
- initializes the cluster weights MMSD_Cluster::setWeightInitializationType set by MMSD_Model::setWeightInitializationType()
- initializes the cluster MMSD_Cluster::initialize()
- if the cluster is not empty, adds it in the list
- verifies the consistency of the cluster:
- the added cluster must not be empty (raises an error if not)
- the parameters of the cluster law must not be out of bounds (write a warning if not)
- copy the initial cluster indices in the mSampleClusterIndices int vector.
- compute the cluster probablity for each sample. (see MMSD_Model::computeSampleClusterProbabilities())
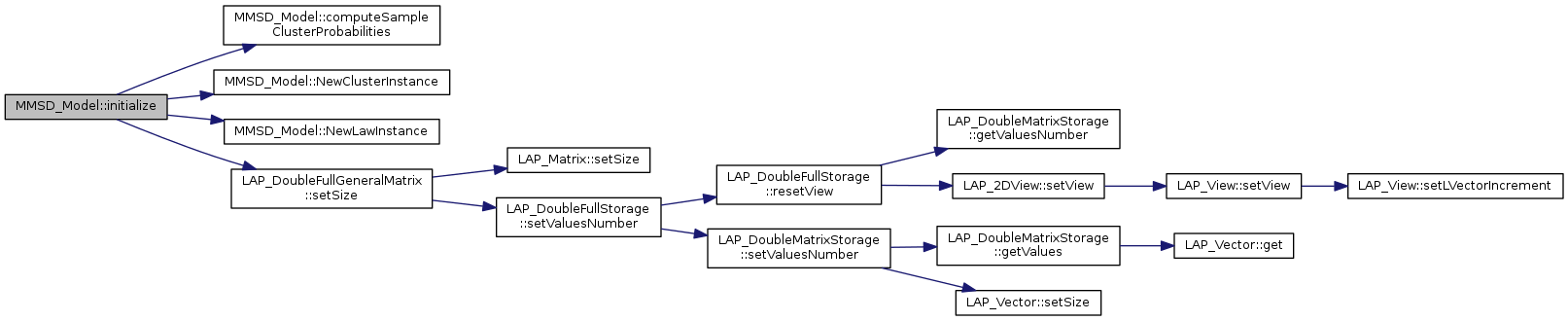
References computeSampleClusterProbabilities(), NewClusterInstance(), NewLawInstance(), LAP_DoubleFullGeneralMatrix::setSize(), tBoolean, and tString.
Referenced by parametersOptimizationByEMMethod().

|
staticinherited |
init the seed with v value
References STAT_Distribution::initSeed().
|
staticinherited |
init the seed
References CORE_Time::getTime().
Referenced by MMSD_Object::MMSD_Object(), testGaussianLaw11N(), testGaussianLaw1PN(), and testGaussianLaw2PN().


|
inlinestaticinherited |
return true if the machine is a 32 bits machine
References CORE_Object::is64Architecture().

|
staticinherited |
return true if the machine is a 64 bits machine
Referenced by CORE_Object::is32Architecture().

|
inlineinherited |
return true if the object is an instance of T
References null.
|
virtual |
how to load the object from a Meta Model class
Reimplemented from UI_Object.
References UI_Class::getInterfaceType(), UI_Class::loadAssociation(), UI_Class::loadField(), LAP_DoubleFullGeneralMatrix::setValues(), LAP_Vector< T >::setValues(), and tBoolean.

|
pure virtual |
create a cluster
Implemented in MMSD_GaussianModel, and MMSD_ConstraintGaussianModel.
Referenced by initialize().

|
pure virtual |
create a cluster
Implemented in MMSD_GaussianModel.
Referenced by initialize().

|
staticinherited |
print on output
References null, and CORE_Object::print().
Referenced by CORE_Exception::CORE_Exception().

| tBoolean MMSD_Model::parametersOptimizationByEMMethod | ( | const int & | nClusters, |
| const MMSD_IntegerVector & | sampleClusters, | ||
| const tString & | backupPath, | ||
| const tString & | backupPrefix, | ||
| const int & | nDigits, | ||
| const int & | backupSteps | ||
| ) |
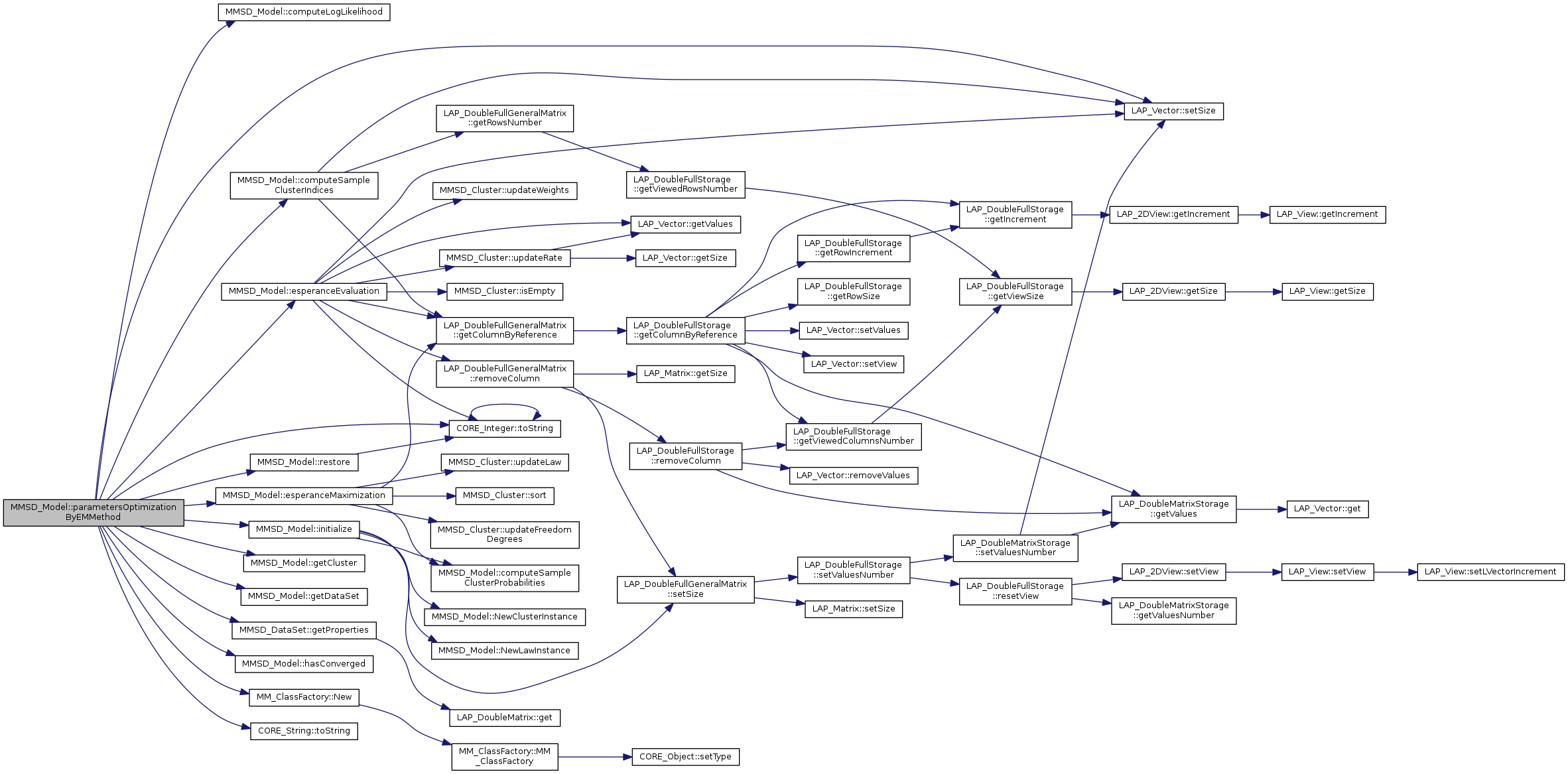
optimise the parameters of each laws by EM algorithm
- Parameters
-
nClusters the number of clusters sampleClusters the cluster index of each sample backupPath : the path to store the results backupPrefix : the prefix of all saved files nDigits the number of print digits for real values backupSteps the step size for saving file
- Returns
- true if the method succeeds.
A data set MMSD_DataSet must be attached to the model. (MMSD_Model::setDataSet). If no data set is provided an exception is raised.
The algorithm is as follow:
- initialization step: if the number of clusters is null, it means that the initialization of the process has to be restored from a preceding computations. (see MMSD_Model::restore() ). Otherwise, the initialize method is invoked (MMSD_Model::initialize())
- while EM iterations number less than max EM iterations number (see MMSD_Model::setEMMaximumIterationsNumber() ) do
- compute esperance (see MMSD_Model::esperanceEvaluation() )
- maximize esperance (see MMSD_Model::esperancemaximization() )
- verify the consistency of the law parameters (see MMSD_Law::isOutOfBounds() )
- compute the log-likelihood & store it (see MMSD_Model::getLogL() )
- verify the log-likelihood is not a real number, in than case raises a nan exception
- save the data for future restore if needed
- compute the sample cluster index (see MMSD_Model::computeSampleClusterIndices())
References computeLogLikelihood(), computeSampleClusterIndices(), esperanceEvaluation(), esperanceMaximization(), getCluster(), getDataSet(), MMSD_DataSet::getProperties(), hasConverged(), initialize(), MM_ClassFactory::New(), null, CORE_File::PATH_SEPARATOR, restore(), LAP_Vector< T >::setSize(), tBoolean, CORE_Integer::toString(), CORE_String::toString(), and tString.
|
inline |
optimise the parameters of each laws by EM algorithm
- Parameters
-
nClusters the number of clusters sampleClusters the cluster index of each sample
- Returns
- true if the method succeeds.
References parametersOptimizationByEMMethod().

|
inline |
optimise the parameters of each laws by EM algorithm
- Returns
- true if the method succeeds.
Referenced by parametersOptimizationByEMMethod().

|
inline |
optimise the parameters of each laws by EM algorithm
- Parameters
-
backupPath : the path to store the results backupPrefix : the prefix of all saved files nDigits the number of print digits for real values backupSteps the step size for saving file
- Returns
- true if the method succeeds.
References parametersOptimizationByEMMethod().

|
staticinherited |
return the string represantation of a pointer
References tString.
Referenced by CORE_Object::CORE_Object(), CORE_Object::getIdentityString(), CORE_Object::getPointerAddress(), and CORE_Object::~CORE_Object().
|
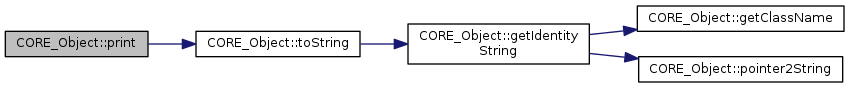
inlinevirtualinherited |
print the class
References CORE_Object::toString().
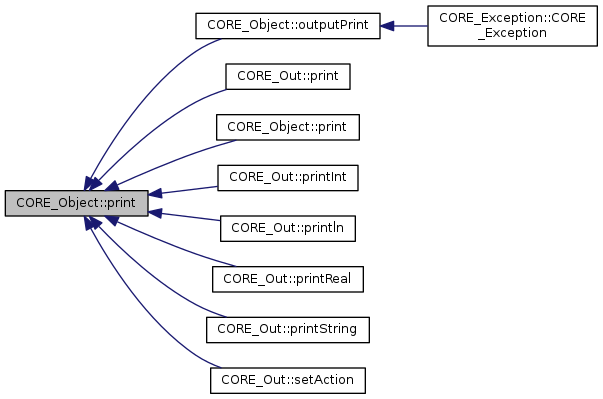
Referenced by CORE_Object::outputPrint(), CORE_Out::print(), CORE_Object::print(), CORE_Out::printInt(), CORE_Out::println(), CORE_Out::printReal(), CORE_Out::printString(), and CORE_Out::setAction().
|
inlinevirtualinherited |
print the class
References CORE_Object::print(), and CORE_Object::toString().

|
virtualinherited |
print the class
Reimplemented in CORE_Out.
References null, and CORE_Object::print().

|
virtualinherited |
References null.
|
virtualinherited |
References null.
|
inlinestaticinherited |
print the class
|
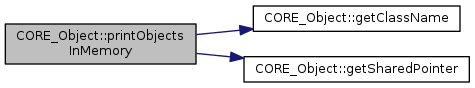
staticinherited |
print object in memory
References CORE_Object::getClassName(), CORE_Object::getSharedPointer(), and CORE_Object::mIsMemoryTesting.
Referenced by main().

|
inline |
restore the model from a file
- Parameters
-
backupPath : the backup path backupPrefix : the backup prefix nDigits the number of print digits for EM iteration step the backup step
- Returns
- true if the restoring succeeds
References CORE_File::PATH_SEPARATOR, CORE_Integer::toString(), and tString.
Referenced by parametersOptimizationByEMMethod().
restore the model from a backup file
- Parameters
-
factory : class factory to generate instances of class file backup file
- Returns
- true if the restoring succeeds
References MM_ClassFactory::New().

|
protectedvirtual |
restore the unstored values after a backup
- Parameters
-
properties the data set properties
- restore all the cluster computing ( see MMSD_Cluster::restore())
- recompute the clusters probablity for each sample. (see MMSD_Model::computeSampleClusterProbabilities())
- update the weight of each cluster thanks to the clusters probability for each sample (see MMSD_Cluster::updateWeights())
References computeSampleClusterProbabilities(), LAP_DoubleFullGeneralMatrix::getColumnByReference(), LAP_DoubleFullGeneralMatrix::getRowsNumber(), and LAP_DoubleFullGeneralMatrix::setSize().
|
virtual |
how to save the object from a mate model class
Reimplemented from UI_Object.
References UI_Class::getInterfaceType(), LAP_Vector< T >::getSize(), LAP_DoubleMatrix::getStorage(), LAP_DoubleMatrixStorage::getValues(), LAP_Vector< T >::getValues(), LAP_DoubleMatrix::getValuesNumber(), UI_Class::saveAssociation(), and UI_Class::saveField().
|
inline |
set the data set
|
inline |
set the maximum iterations number for EM algorithm
see MMSD_Model::
|
inlineinherited |
set the if the object has completely been loaded
Referenced by UI_Class::loadAssociation().

|
inline |
if true the empty clusters are deleted during esperaceEvaluation() defalut value: false. see MMSD_Model:::esperanceEvaluation()
|
inline |
set min eigen value for eigen value decomposition of property covariance matrix law
|
inlinestaticinherited |
set output
|
inlineprotectedinherited |
set this weak shared pointer called toDoAfterThis setting method
References CORE_Object::toDoAfterThisSetting().

|
inlineprotectedvirtualinherited |
set the type of the object
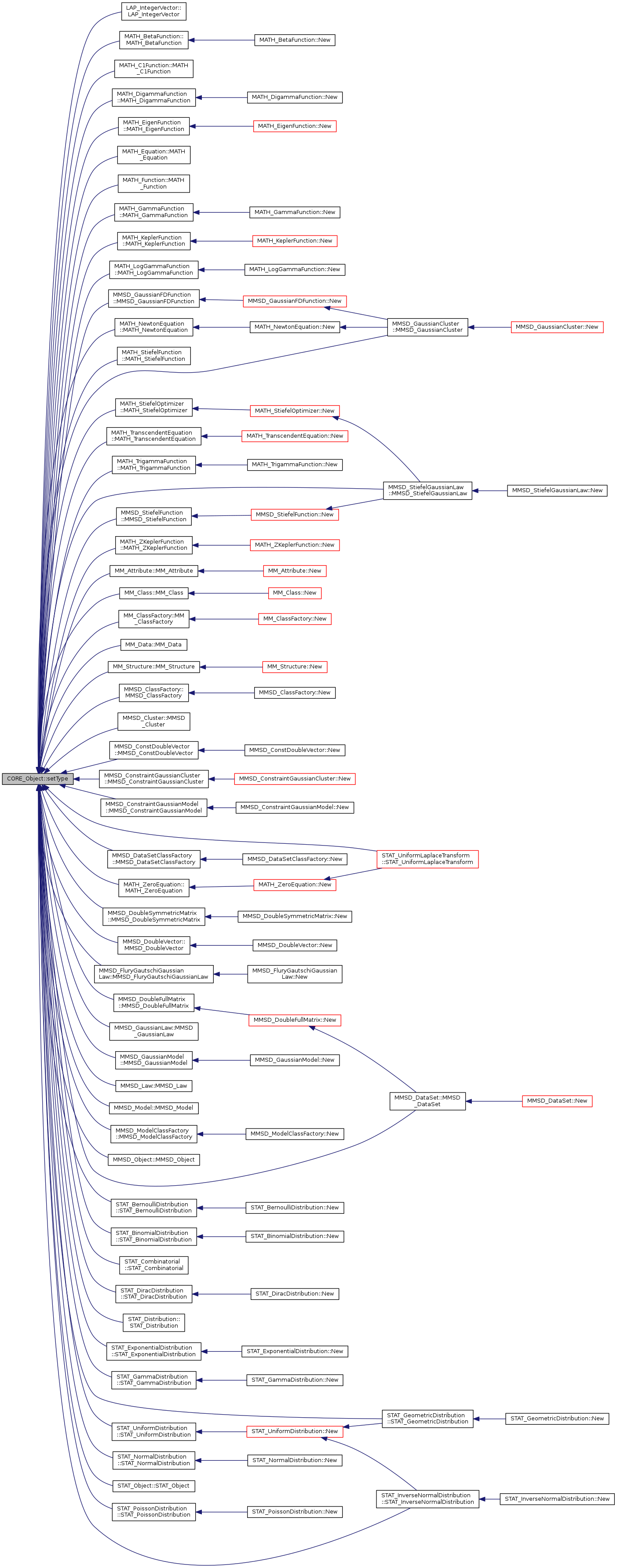
Referenced by LAP_IntegerVector::LAP_IntegerVector(), MATH_BetaFunction::MATH_BetaFunction(), MATH_C1Function::MATH_C1Function(), MATH_DigammaFunction::MATH_DigammaFunction(), MATH_EigenFunction::MATH_EigenFunction(), MATH_Equation::MATH_Equation(), MATH_Function::MATH_Function(), MATH_GammaFunction::MATH_GammaFunction(), MATH_KeplerFunction::MATH_KeplerFunction(), MATH_LogGammaFunction::MATH_LogGammaFunction(), MATH_NewtonEquation::MATH_NewtonEquation(), MATH_StiefelFunction::MATH_StiefelFunction(), MATH_StiefelOptimizer::MATH_StiefelOptimizer(), MATH_TranscendentEquation::MATH_TranscendentEquation(), MATH_TrigammaFunction::MATH_TrigammaFunction(), MATH_ZeroEquation::MATH_ZeroEquation(), MATH_ZKeplerFunction::MATH_ZKeplerFunction(), MM_Attribute::MM_Attribute(), MM_Class::MM_Class(), MM_ClassFactory::MM_ClassFactory(), MM_Data::MM_Data(), MM_Structure::MM_Structure(), MMSD_ClassFactory::MMSD_ClassFactory(), MMSD_Cluster::MMSD_Cluster(), MMSD_ConstDoubleVector::MMSD_ConstDoubleVector(), MMSD_ConstraintGaussianCluster::MMSD_ConstraintGaussianCluster(), MMSD_ConstraintGaussianModel::MMSD_ConstraintGaussianModel(), MMSD_DataSet::MMSD_DataSet(), MMSD_DataSetClassFactory::MMSD_DataSetClassFactory(), MMSD_DoubleFullMatrix::MMSD_DoubleFullMatrix(), MMSD_DoubleSymmetricMatrix::MMSD_DoubleSymmetricMatrix(), MMSD_DoubleVector::MMSD_DoubleVector(), MMSD_FluryGautschiGaussianLaw::MMSD_FluryGautschiGaussianLaw(), MMSD_GaussianCluster::MMSD_GaussianCluster(), MMSD_GaussianFDFunction::MMSD_GaussianFDFunction(), MMSD_GaussianLaw::MMSD_GaussianLaw(), MMSD_GaussianModel::MMSD_GaussianModel(), MMSD_Law::MMSD_Law(), MMSD_Model(), MMSD_ModelClassFactory::MMSD_ModelClassFactory(), MMSD_Object::MMSD_Object(), MMSD_StiefelFunction::MMSD_StiefelFunction(), MMSD_StiefelGaussianLaw::MMSD_StiefelGaussianLaw(), STAT_BernoulliDistribution::STAT_BernoulliDistribution(), STAT_BinomialDistribution::STAT_BinomialDistribution(), STAT_Combinatorial< T >::STAT_Combinatorial(), STAT_DiracDistribution::STAT_DiracDistribution(), STAT_Distribution::STAT_Distribution(), STAT_ExponentialDistribution::STAT_ExponentialDistribution(), STAT_GammaDistribution::STAT_GammaDistribution(), STAT_GeometricDistribution::STAT_GeometricDistribution(), STAT_InverseNormalDistribution::STAT_InverseNormalDistribution(), STAT_NormalDistribution::STAT_NormalDistribution(), STAT_Object::STAT_Object(), STAT_PoissonDistribution::STAT_PoissonDistribution(), STAT_UniformDistribution::STAT_UniformDistribution(), and STAT_UniformLaplaceTransform::STAT_UniformLaplaceTransform().
|
inline |
set the weight initialization type & parameters
- Parameters
-
type of weight initialization ["gamma" or "constant"] shape : parameter for statistic function weight initialization rate : parameter for statistic function weight initialization
|
inlineprotectedvirtualinherited |
method called after setThis() method this method can oly be called once.
Reimplemented in STAT_UniformLaplaceTransform.
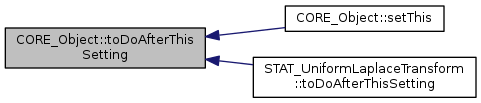
Referenced by CORE_Object::setThis(), and STAT_UniformLaplaceTransform::toDoAfterThisSetting().
|
inlinevirtual |
turn the class into string
Reimplemented from CORE_Object.
References CORE_Object::toString(), and tString.

Member Data Documentation
|
staticinherited |
indicator to store all classes created and deleted only for debuging version
Referenced by CORE_Object::CORE_Object(), main(), CORE_Object::printObjectsInMemory(), and CORE_Object::~CORE_Object().
The documentation for this class was generated from the following files:

 1.8.8
1.8.8