The purpose of this package is to determine the group of each sample of a set of samples with properties.
A data set MMSD_DataSet contains a properties matrix of size NxP.
Initialy a group is attached to each samples.
Thanks to an implementation of model class MMSD_Model , an optimization is done to compute the best group attached to each sample by the method MMSD_Model::parametersOptimizationByEMMethod see MMSD_Core package.
The model implementations are done in the package MMSD_Model
An example is given by A. Arnaud applied to the MRI for rats brain.
The data set is the sequence of MRI of size N with following properties:
- ADC (Apparent diffusion coefficient; m^2/s)
- AUC (Blood vessel permeability; U.A)
- CBV (cerebral blood volume; %)
- T1 (spin-lattice relaxation time; s)
- T2 (spin-spin relaxation time; s)
The first step is to clustering the MRI in 11 healthy classes. At each set of voxel is associated an (more or less) arbitraty class and then, thanks to the optimal process, an optimal healthy model is detected:
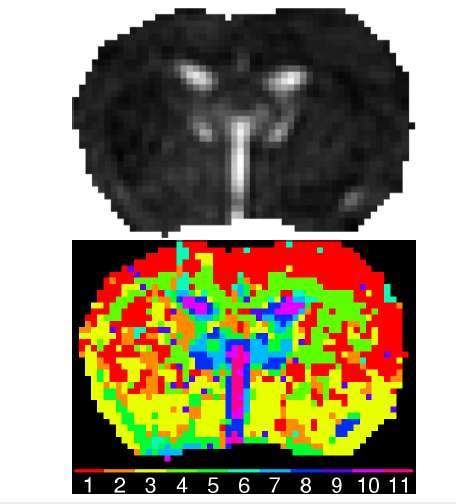
.
The second steps consist in finding the outlier voxels of MRI of sick rats thanks to an outlierness threshold. Then only this atypical data is focused on the last steps.
The last steps is to clustering the outlier area of sick MRI in sick classes like the first steps.
Thanks to this 3 proceses, it is possible to make a map for each considering tumor (9L, C6,F98,RG2) or healthy depending on the outlerness threshold:
From the preceeding maps, a tumors dictionary can be build:
For each new sick rat, a clustering is done and the results is compared with the dictionay to detect its tumors.
